Interpretable End-to-End Learning
Sheffield ML Group Research Retreat
Bits and Atoms
Supply Chain

Cromford


Deep Freeze

Deep Freeze

Machine Learning in Supply Chain
- Supply chain: Large Automated Decision Making Network
- Major Challenge:
- We have a mechanistic understanding of supply chain.
- Machine learning is a data driven technology.
Motto
Solve Supply Chain, then solve everything else.
From Model to Decision

|
\[\text{data} + \text{model} \xrightarrow{\text{compute}} \text{prediction}\] |
Experiment, Analyze, Design
Our Vision
We don’t know what science we’ll want to do in 5 years time, but we won’t want slower experiments, we won’t want more expensive experiments and we won’t want a narrower selection of experiments.
What do we want?
- Faster, cheaper and more diverse experiments.
- Better ecosystems for experimentation.
- Data oriented architectures.
Data Oriented Architectures
- Convert data to a first-class citizen.
- View system as operations on data streams.
- Expose data operations in a programmatic way.
Data Orientated Architectures
- Historically we’ve been software first
- A necessary but not sufficient condition for data first
- Move from
- service orientated architectures
- data orientated architectures
Streaming System
- Move from pull updates to push updates.
- Operate on rows rather than columns.
- Lead to stateless logic: persistence handled by system.
- Example Apache Kafka + Apache Flink
Streaming Architectures
- AWS Kinesis, Apache Kafka
- Not just about streaming
- Nodes in the architecture are stateless
- They persist through storing state on streams
- This brings the data inside out
Apache Flink
- Streams and transformations
- a stream is a (potentially never-ending) flow of data records
- a transformation: streams as input, produces transformed streams as output
Join
stream.join(otherStream)
.where(<KeySelector>)
.equalTo(<KeySelector>)
.window(<WindowAssigner>)
.apply(<JoinFunction>)Milan
- Data Oriented Programming Language and runtime.
- DSL Embedded in Scala converts to an intermediate langugage.
- Intermediate language for compilation on different platforms (currently Flink)
Trading System
- High frequency share trading.
- Stream of prices with millisecond updates.
- Trades required on millisecond time line
Real Price
Future Price
Hypothetical Streams
- Real stream — share prices
- derived hypothetical stream — share prices in future.
- Hypothetical constrained by
- input constraints.
- decision functional
- computational requirements (latency)
Hypothetical Advantage
- Modelling is now required.
- But modelling is declared in the ecosystem.
- If it’s manual, warnings can be used
- calibration, fairness, dataset shift
- Opens door to Auto AI.
SafeBoda
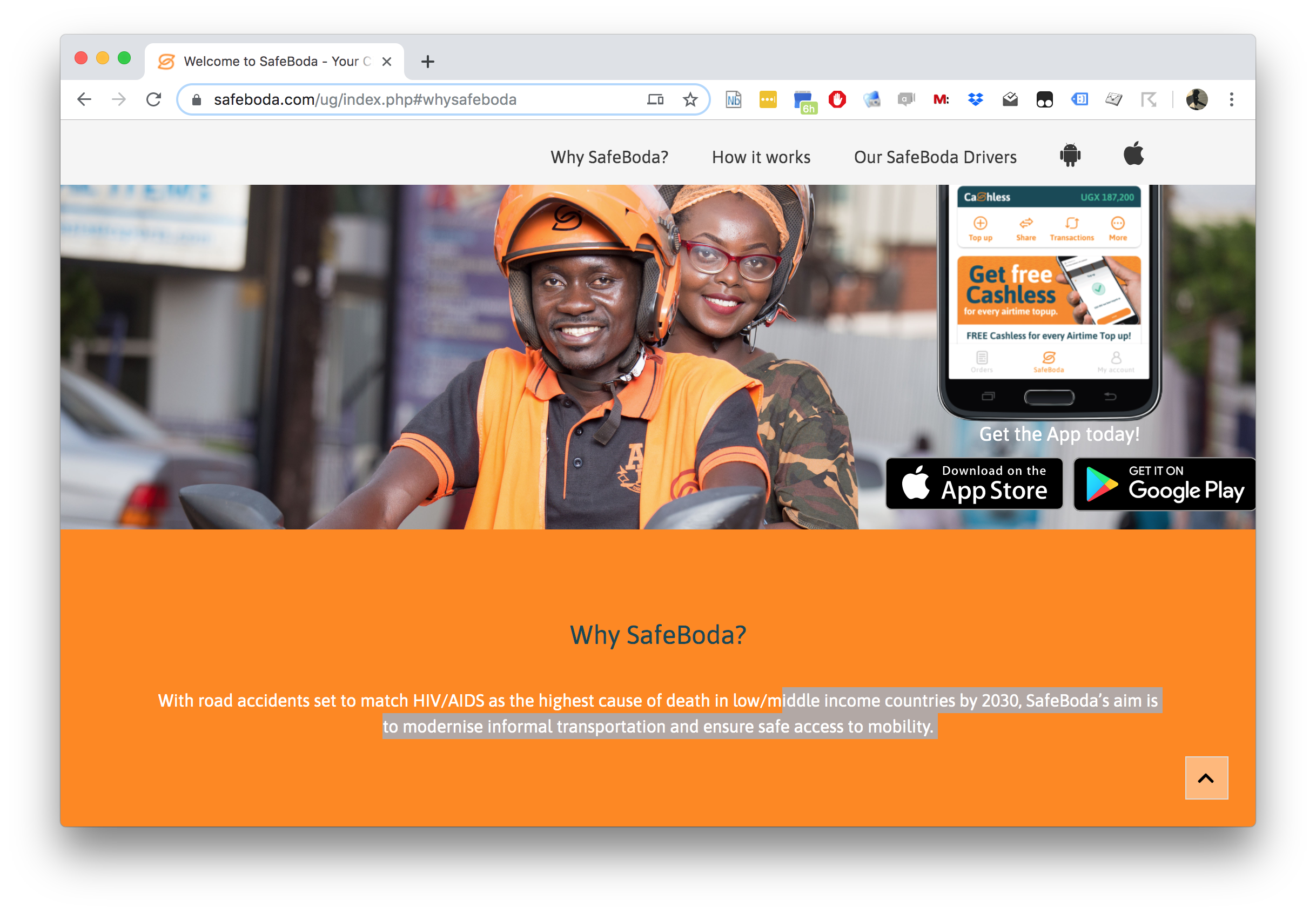
SafeBoda
With road accidents set to match HIV/AIDS as the highest cause of death in low/middle income countries by 2030, SafeBoda’s aim is to modernise informal transportation and ensure safe access to mobility.
Ride Allocation Prediction
Ride Sharing: Service Oriented
Ride Sharing: Data Oriented
Ride Sharing: Hypothetical
Information Dynamics
- Potential for information feedback loops.
- Hypothetical streams are instantiated.
- Nature hypothesis (e.g. price prediction) can effect reality.
- Leads to information dynamics, similar to dynamics of governors.
- See e.g. Closed Loop Data Science at Glasgow.
Autonomous Vehicles
Emulation
Emulation
Emulation
Emulation
Emulation
Emulation
Deep Emulation
Deep Emulation
Deep Emulation
Bayesian System Optimization
- Aim: maintain interpretable components.
- Monitor downstream/upstream effects through emulation.
- Optimize individual components considering upstream and downstream.
Auto AI
- Auto ML is great but not sufficient
- Interacting components in an ML system
- Identify problems, and automatically deploy solutions
Technology: Deep Emulation
DeepFace
Outline of the DeepFace architecture. A front-end of a single convolution-pooling-convolution filtering on the rectified input, followed by three locally-connected layers and two fully-connected layers. Color illustrates feature maps produced at each layer. The net includes more than 120 million parameters, where more than 95% come from the local and fully connected.
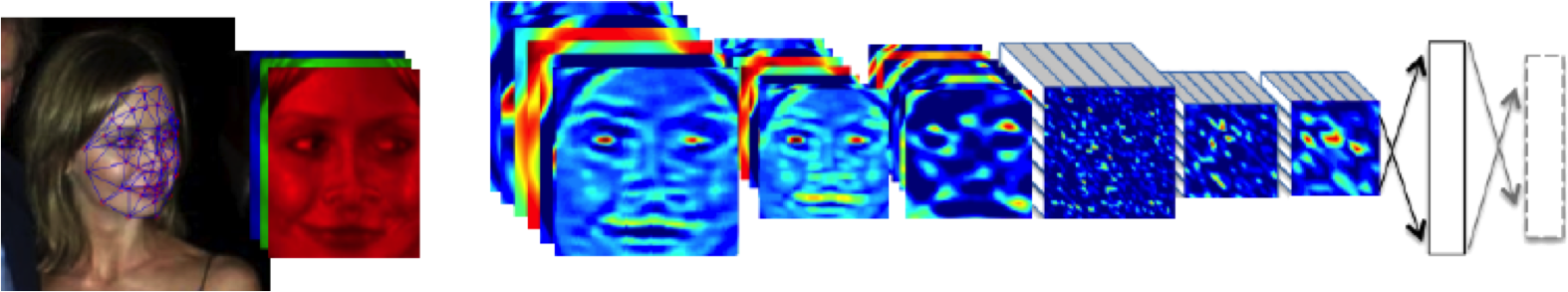
Source: DeepFace (Taigman et al., 2014)
Deep Neural Network
Deep Neural Network
Mathematically
\[ \begin{align} \hiddenVector_{1} &= \basisFunction\left(\mappingMatrix_1 \inputVector\right)\\ \hiddenVector_{2} &= \basisFunction\left(\mappingMatrix_2\hiddenVector_{1}\right)\\ \hiddenVector_{3} &= \basisFunction\left(\mappingMatrix_3 \hiddenVector_{2}\right)\\ \dataVector &= \mappingVector_4 ^\top\hiddenVector_{3} \end{align} \]
Overfitting
Potential problem: if number of nodes in two adjacent layers is big, corresponding \(\mappingMatrix\) is also very big and there is the potential to overfit.
Proposed solution: “dropout”.
Alternative solution: parameterize \(\mappingMatrix\) with its SVD. \[ \mappingMatrix = \eigenvectorMatrix\eigenvalueMatrix\eigenvectwoMatrix^\top \] or \[ \mappingMatrix = \eigenvectorMatrix\eigenvectwoMatrix^\top \] where if \(\mappingMatrix \in \Re^{k_1\times k_2}\) then \(\eigenvectorMatrix\in \Re^{k_1\times q}\) and \(\eigenvectwoMatrix \in \Re^{k_2\times q}\), i.e. we have a low rank matrix factorization for the weights.
Low Rank Approximation
Bottleneck Layers in Deep Neural Networks
Deep Neural Network
Mathematically
The network can now be written mathematically as \[ \begin{align} \latentVector_{1} &= \eigenvectwoMatrix^\top_1 \inputVector\\ \hiddenVector_{1} &= \basisFunction\left(\eigenvectorMatrix_1 \latentVector_{1}\right)\\ \latentVector_{2} &= \eigenvectwoMatrix^\top_2 \hiddenVector_{1}\\ \hiddenVector_{2} &= \basisFunction\left(\eigenvectorMatrix_2 \latentVector_{2}\right)\\ \latentVector_{3} &= \eigenvectwoMatrix^\top_3 \hiddenVector_{2}\\ \hiddenVector_{3} &= \basisFunction\left(\eigenvectorMatrix_3 \latentVector_{3}\right)\\ \dataVector &= \mappingVector_4^\top\hiddenVector_{3}. \end{align} \]
A Cascade of Neural Networks
\[ \begin{align} \latentVector_{1} &= \eigenvectwoMatrix^\top_1 \inputVector\\ \latentVector_{2} &= \eigenvectwoMatrix^\top_2 \basisFunction\left(\eigenvectorMatrix_1 \latentVector_{1}\right)\\ \latentVector_{3} &= \eigenvectwoMatrix^\top_3 \basisFunction\left(\eigenvectorMatrix_2 \latentVector_{2}\right)\\ \dataVector &= \mappingVector_4 ^\top \latentVector_{3} \end{align} \]
Cascade of Gaussian Processes
Replace each neural network with a Gaussian process \[ \begin{align} \latentVector_{1} &= \mappingFunctionVector_1\left(\inputVector\right)\\ \latentVector_{2} &= \mappingFunctionVector_2\left(\latentVector_{1}\right)\\ \latentVector_{3} &= \mappingFunctionVector_3\left(\latentVector_{2}\right)\\ \dataVector &= \mappingFunctionVector_4\left(\latentVector_{3}\right) \end{align} \]
Equivalent to prior over parameters, take width of each layer to infinity.
Stochastic Process Composition
\[\dataVector = \mappingFunctionVector_4\left(\mappingFunctionVector_3\left(\mappingFunctionVector_2\left(\mappingFunctionVector_1\left(\inputVector\right)\right)\right)\right)\]
Motorcycle Helmet Data
Motorcycle Helmet Data GP
Motorcycle Helmet Data Deep GP
Motorcycle Helmet Data Deep GP
Motorcycle Helmet Data Latent 1
Motorcycle Helmet Data Latent 2
Motorcycle Helmet Pinball Plot
Graphical Models
- Represent joint distribution through conditional dependencies.
- E.g. Markov chain
\[p(\dataVector) = p(\dataScalar_\numData | \dataScalar_{\numData-1}) p(\dataScalar_{\numData-1}|\dataScalar_{\numData-2}) \dots p(\dataScalar_{2} | \dataScalar_{1})\]
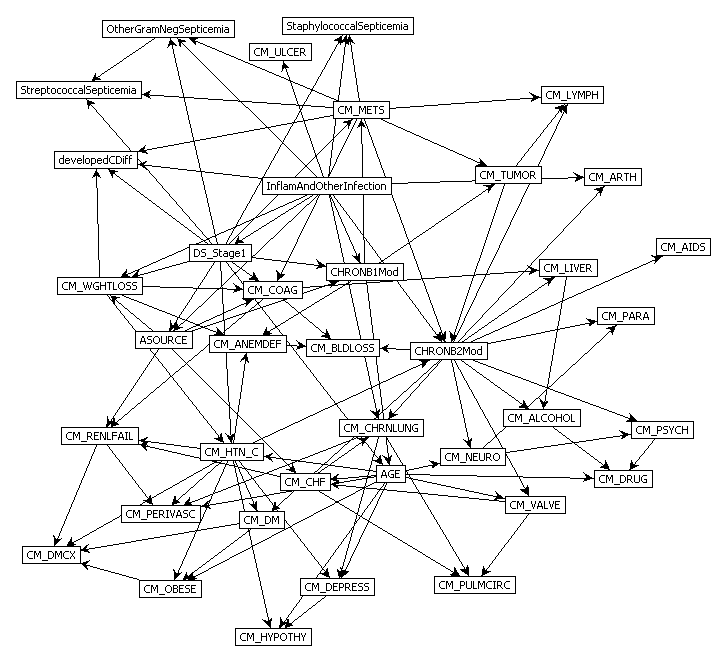
Predict Perioperative Risk of Clostridium Difficile Infection Following Colon Surgery (Steele et al., 2012)
Conclusion
- Challenges in decomposition, data and model deployment for ML.
- Data oriented architectures and data first thinking are the solution.
- Data oriented programming creates systems that are ready to deploy.
- Opens the door to AutoAI and information dynamics analysis.
Related Papers
Deep Gaussian Processes Damianou and Lawrence (2013)
Latent Force Models Álvarez et al. (2013)
Gaussian Process Latent Force Models for Learning and Stochastic Control of Physical Systems Särkkä et al. (2018)
The Emergence of Organizing Structure in Conceptual Representation Lake et al. (2018)
Other’s Work
- How Deep Are Deep Gaussian Processes? Dunlop et al. (n.d.)
- Doubly Stochastic Variational Inference for Deep Gaussian Processes Salimbeni and Deisenroth (2017)
- Deep Multi-task Gaussian Processes for Survival Analysis with Competing Risks Alaa and van der Schaar (2017)
- Counterfactual Gaussian Processes for Reliable Decision-making and What-if Reasoning Schulam and Saria (2017)
Conclusions and Directions
- Mechanistic modelling
- Automated Abstraction
- Deep emulation
- Bayesian Systems Optimization
- Auto AI
Thanks!
- twitter: @lawrennd
- podcast: The Talking Machines
- newspaper: Guardian Profile Page
- blog: http://inverseprobability.com
References
Alaa, A.M., van der Schaar, M., 2017. Deep multi-task Gaussian processes for survival analysis with competing risks, in: Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R. (Eds.), Advances in Neural Information Processing Systems 30. Curran Associates, Inc., pp. 2326–2334.
Álvarez, M.A., Luengo, D., Lawrence, N.D., 2013. Linear latent force models using Gaussian processes. IEEE Transactions on Pattern Analysis and Machine Intelligence 35, 2693–2705. https://doi.org/10.1109/TPAMI.2013.86
Damianou, A., Lawrence, N.D., 2013. Deep Gaussian processes, in:. pp. 207–215.
Dunlop, M.M., Girolami, M.A., Stuart, A.M., Teckentrup, A.L., n.d. How deep are deep Gaussian processes? Journal of Machine Learning Research 19, 1–46.
Lake, B.M., Lawrence, N.D., Tenenbaum, J.B., 2018. The emergence of organizing structure in conceptual representation. Cognitive science 42 Suppl 3, 809–832. https://doi.org/10.1111/cogs.12580
Salimbeni, H., Deisenroth, M., 2017. Doubly stochastic variational inference for deep Gaussian processes, in: Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R. (Eds.), Advances in Neural Information Processing Systems 30. Curran Associates, Inc., pp. 4591–4602.
Särkkä, S., Álvarez, M.A., Lawrence, N.D., 2018. Gaussian process latent force models for learning and stochastic control of physical systems. IEEE Transactions on Automatic Control. https://doi.org/10.1109/TAC.2018.2874749
Schulam, P., Saria, S., 2017. Counterfactual Gaussian processes for reliable decision-making and what-if reasoning, in: Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R. (Eds.), Advances in Neural Information Processing Systems 30. Curran Associates, Inc., pp. 1696–1706.
Steele, S., Bilchik, A., Eberhardt, J., Kalina, P., Nissan, A., Johnson, E., Avital, I., Stojadinovic, A., 2012. Using machine-learned Bayesian belief networks to predict perioperative risk of clostridium difficile infection following colon surgery. Interact J Med Res 1, e6. https://doi.org/10.2196/ijmr.2131
Taigman, Y., Yang, M., Ranzato, M., Wolf, L., 2014. DeepFace: Closing the gap to human-level performance in face verification, in: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. https://doi.org/10.1109/CVPR.2014.220