Towards Machine Learning Systems Design
Department of Computing Science, University of Glasgow
What is Machine Learning?
\[ \text{data} + \text{model} \xrightarrow{\text{compute}} \text{prediction}\]
- data : observations, could be actively or passively acquired (meta-data).
- model : assumptions, based on previous experience (other data! transfer learning etc), or beliefs about the regularities of the universe. Inductive bias.
- prediction : an action to be taken or a categorization or a quality score.
- Royal Society Report: Machine Learning: Power and Promise of Computers that Learn by Example
What is Machine Learning?
\[\text{data} + \text{model} \xrightarrow{\text{compute}} \text{prediction}\]
- To combine data with a model need:
- a prediction function \(\mappingFunction (\cdot)\) includes our beliefs about the regularities of the universe
- an objective function \(\errorFunction (\cdot)\) defines the cost of misprediction.
Machine Learning
- Driver of two different domains:
- Data Science: arises from the fact that we now capture data by happenstance.
- Artificial Intelligence: emulation of human behaviour.
- Connection: Internet of Things
Machine Learning
- Driver of two different domains:
- Data Science: arises from the fact that we now capture data by happenstance.
- Artificial Intelligence: emulation of human behaviour.
- Connection: Internet of
Things
Machine Learning
- Driver of two different domains:
- Data Science: arises from the fact that we now capture data by happenstance.
- Artificial Intelligence: emulation of human behaviour.
- Connection: Internet of People
What does Machine Learning do?
- ML Automates through Data
- Strongly related to statistics.
- Field underpins revolution in data science and AI
- With AI:
- logic, robotics, computer vision, speech
- With Data Science:
- databases, data mining, statistics, visualization
Embodiment Factors

|

|
|
| compute | \[\approx 100 \text{ gigaflops}\] | \[\approx 16 \text{ petaflops}\] |
| communicate | \[1 \text{ gigbit/s}\] | \[100 \text{ bit/s}\] |
| (compute/communicate) | \[10^{4}\] | \[10^{14}\] |
See “Living Together: Mind and Machine Intelligence” Lawrence (2017)



.
Evolved Relationship
Evolved Relationship
What does Machine Learning do?
- Automation scales by codifying processes and automating them.
- Need:
- Interconnected components
- Compatible components
- Early examples:
- cf Colt 45, Ford Model T
Codify Through Mathematical Functions
- How does machine learning work?
- Jumper (jersey/sweater) purchase with logistic regression
\[ \text{odds} = \frac{p(\text{bought})}{p(\text{not bought})} \]
\[ \log \text{odds} = \beta_0 + \beta_1 \text{age} + \beta_2 \text{latitude}.\]
Codify Through Mathematical Functions
- How does machine learning work?
- Jumper (jersey/sweater) purchase with logistic regression
\[ p(\text{bought}) = \sigmoid{\beta_0 + \beta_1 \text{age} + \beta_2 \text{latitude}}.\]
Codify Through Mathematical Functions
- How does machine learning work?
- Jumper (jersey/sweater) purchase with logistic regression
\[ p(\text{bought}) = \sigmoid{\boldsymbol{\beta}^\top \inputVector}.\]
Codify Through Mathematical Functions
- How does machine learning work?
- Jumper (jersey/sweater) purchase with logistic regression
\[ \dataScalar = \mappingFunction\left(\inputVector, \boldsymbol{\beta}\right).\]
We call \(\mappingFunction(\cdot)\) the prediction function.
Fit to Data
- Use an objective function
\[\errorFunction(\boldsymbol{\beta}, \dataMatrix, \inputMatrix)\]
- E.g. least squares \[\errorFunction(\boldsymbol{\beta}, \dataMatrix, \inputMatrix) = \sum_{i=1}^\numData \left(\dataScalar_i - \mappingFunction(\inputVector_i, \boldsymbol{\beta})\right)^2.\]
Two Components
- Prediction function, \(\mappingFunction(\cdot)\)
- Objective function, \(\errorFunction(\cdot)\)
Deep Learning
These are interpretable models: vital for disease modeling etc.
Modern machine learning methods are less interpretable
Example: face recognition
DeepFace
Outline of the DeepFace architecture. A front-end of a single convolution-pooling-convolution filtering on the rectified input, followed by three locally-connected layers and two fully-connected layers. Color illustrates feature maps produced at each layer. The net includes more than 120 million parameters, where more than 95% come from the local and fully connected.
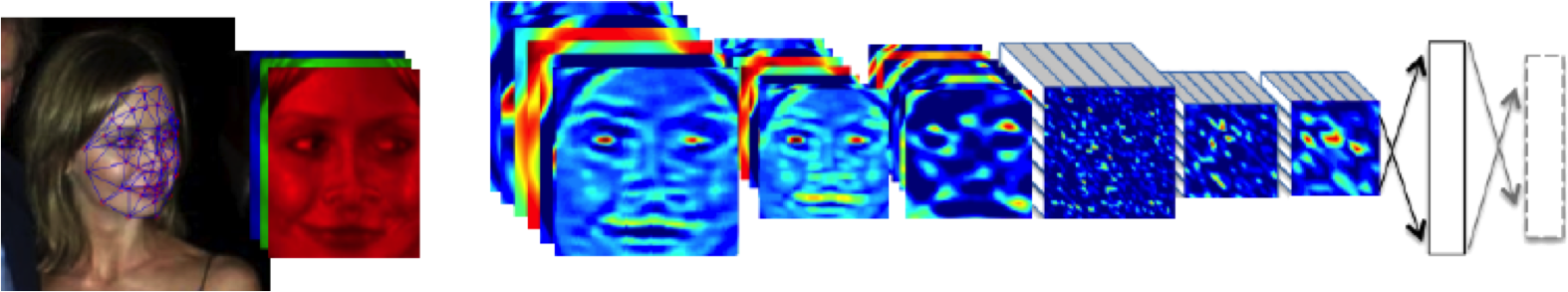
Source: DeepFace (Taigman et al., 2014)
Deep Learning as Pinball



Olympic Marathon Data
|

|
Olympic Marathon Data
Alan Turing

|

|
Probability Winning Olympics?
- He was a formidable Marathon runner.
- In 1946 he ran a time 2 hours 46 minutes.
- That’s a pace of 3.95 min/km.
- What is the probability he would have won an Olympics if one had been held in 1946?
Olympic Marathon Data GP
Deep GP Fit
Can a Deep Gaussian process help?
Deep GP is one GP feeding into another.
Olympic Marathon Data Deep GP
Olympic Marathon Data Deep GP
Olympic Marathon Data Latent 1
Olympic Marathon Data Latent 2
Olympic Marathon Pinball Plot
Supply Chain

Cromford


Deep Freeze

Deep Freeze

Machine Learning in Supply Chain
- Supply chain: Large Automated Decision Making Network
- Major Challenge:
- We have a mechanistic understanding of supply chain.
- Machine learning is a data driven technology.
Deploying Artificial Intelligence
- Challenges in deploying AI.
- Currently this is in the form of “machine learning systems”
Internet of People
- Fog computing: barrier between cloud and device blurring.
- Computing on the Edge
- Complex feedback between algorithm and implementation
Deploying ML in Real World: Machine Learning Systems Design
- Major new challenge for systems designers.
- Internet of Intelligence but currently:
- AI systems are fragile
Machine Learning Systems Design
Fragility of AI Systems
- They are componentwise built from ML Capabilities.
- Each capability is independently constructed and verified.
- Pedestrian detection
- Road line detection
- Important for verification purposes.
Pigeonholing

Robust
- Need to move beyond pigeonholing tasks.
- Need new approaches to both the design of the individual components, and the combination of components within our AI systems.
Rapid Reimplementation
- Whole systems are being deployed.
- But they change their environment.
- The experience evolved adversarial behaviour.
Machine Learning Systems Design

Adversaries
- Stuxnet
- Mischevious-Adversarial
An Intelligent System
Joint work with M. Milo
An Intelligent System
Joint work with M. Milo
Peppercorns
- A new name for system failures which aren’t bugs.
- Difference between finding a fly in your soup vs a peppercorn in your soup.
Peppercorns
Turnaround And Update
- There is a massive need for turn around and update
- A redeploy of the entire system.
- This involves changing the way we design and deploy.
- Interface between security engineering and machine learning.
Emukit
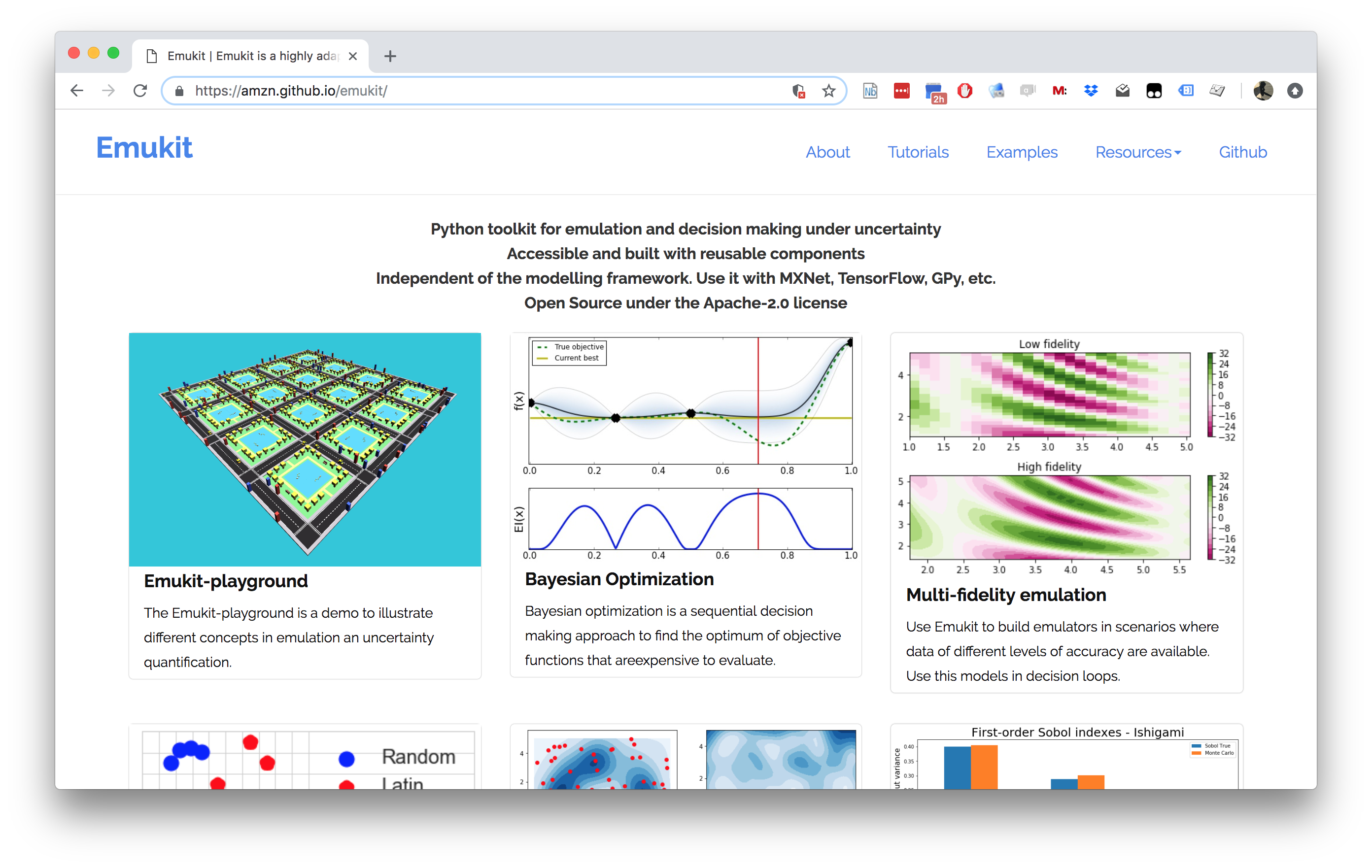
Emukit
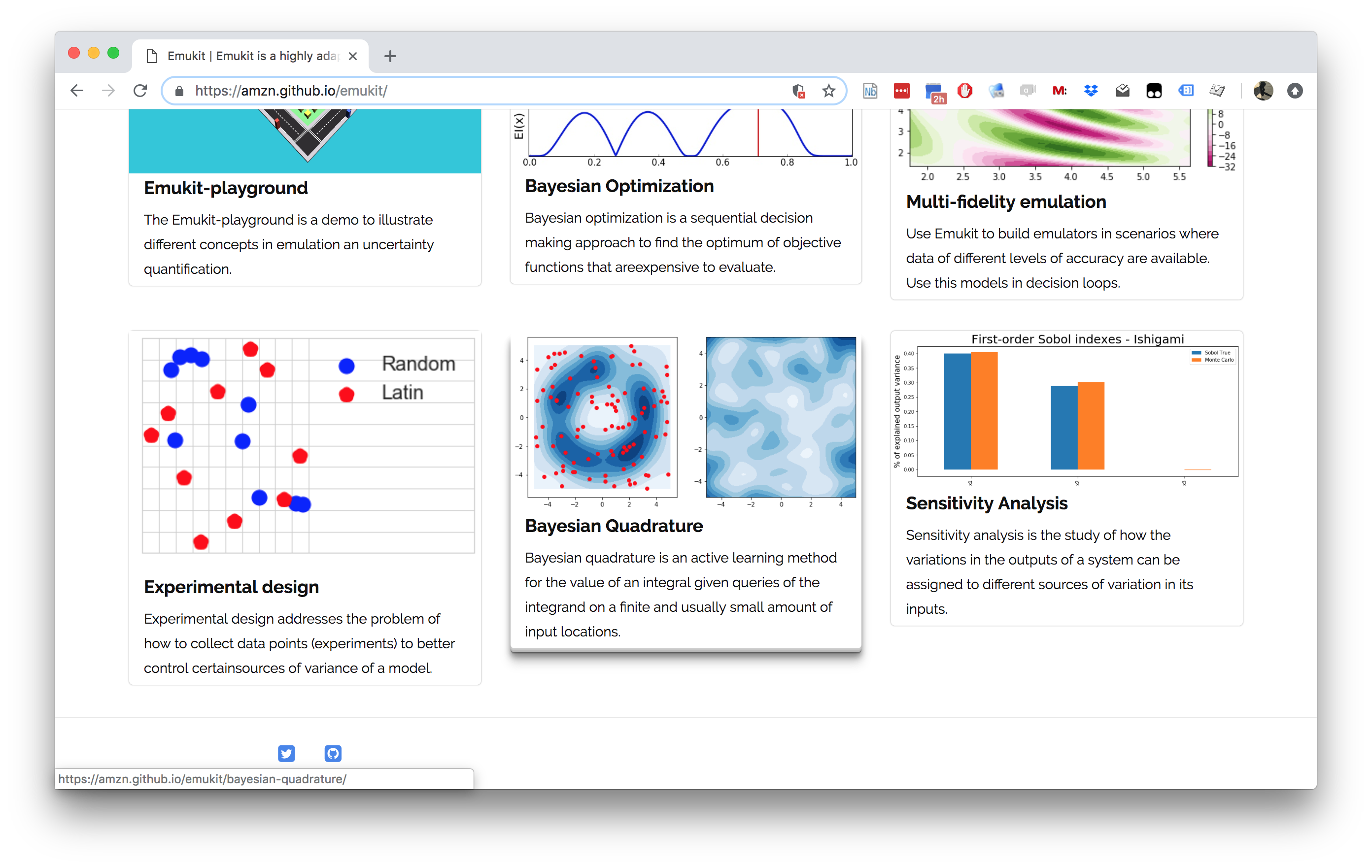
Emukit
- Work by Javier Gonzalez, Andrei Paleyes, Mark Pullin, Maren Mahsereci, Alex Gessner, Aaron Klein.
- Available on Github
- Example sensitivity notebook.
Emukit Software
- Multi-fidelity emulation: build surrogate models for multiple sources of information;
- Bayesian optimisation: optimise physical experiments and tune parameters ML algorithms;
- Experimental design/Active learning: design experiments and perform active learning with ML models;
- Sensitivity analysis: analyse the influence of inputs on the outputs
- Bayesian quadrature: compute integrals of functions that are expensive to evaluate.
Conclusion
Artificial Intelligence and Data Science are fundamentally different.
In one you are dealing with data collected by happenstance.
In the other you are trying to build systems in the real world, often by actively collecting data.
Our approaches to systems design are building powerful machines that will be deployed in evolving environments.
Thanks!
- twitter: @lawrennd
- podcast: The Talking Machines
newspaper: Guardian Profile Page
Blog post on What is Machine Learning?
Blog post on System Zero
Blog post on Decision Making and Diversity
Blog post on Natural vs Artifical Intelligence
References
Lawrence, N.D., 2017. Living together: Mind and machine intelligence. arXiv.
Taigman, Y., Yang, M., Ranzato, M., Wolf, L., 2014. DeepFace: Closing the gap to human-level performance in face verification, in: Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. https://doi.org/10.1109/CVPR.2014.220